NIST 11 (EPA/EPA/NIH Mass Spectral Database of Electron Ionization Data, GC Method/Retention Index Databases, A Database of Spectra Obtained Using MS/MS Techniques, v.2.0g of the NIST MS Search Program, MS Interpreter v.2, and AMDIS)
What’s New and What Value Does it Offer? Part 2
The NIST 11 (National Institute of Standards and Technology) Mass Spectral database, the successor to the NIST 08, is a fully evaluated collection of electron ionization (EI) Mass Spectra, which also includes a growing number of MS/MS Spectra and GC data. In this multi-part article, David Sparkman looks at history and current status of NIST 11 and explains its value to analytical scientists.
In 1971, shortly after the acquisition of its initial six GC/MS instruments, the newly formed United States Government’s Environmental Protection Agency (EPA) awarded a development contract to Battelle-Memorial Institute in Columbus, Ohio, to create a system that automatically transmitted data over voice-grade telephone lines directly from the minicomputers connected to the GC/MS instruments in field laboratories to a program running on a large-scale remote time-sharing computer where the actual search was done against the m/z value and intensity pairs stored as fields in each individual spectrum which constituted records in the database. The database of electron ionization (EI) spectra stored on that time-sharing computer has grown and evolved into what is today’s NIST 11 Database. The evolution of this Database has also lead to the most widely used mass spectral program, the NIST Mass Spectral (MS) Search Program .
NIST Mass Spectral Search Program v.2.0g used with NIST 11
This version of the NIST MS Search Program has many new features that make it easier and more efficient to identify compounds using the mass spectral data provided with the release. As seen in Figure 1, the Program consists of six Tab views, selectable along the bottom of the display. Each view can be brought into focus by either clicking on the Tab or using the Ctrl-Tab and Ctrl-Shift-Tab Hot keys. When the Program is exited and restarted, the last Tab view in focus is brought back into focus. Each Tab view can be configured individually with respect to Windows opens, slider bar positions and orientations, fonts, colors of text, graphics and backgrounds, and the information displayed in the various Windows; i.e., bar graph spectra with or without structures. If multiple people are using the Program , each can have an individual set of configurations because the configurations can be saved and recalled.
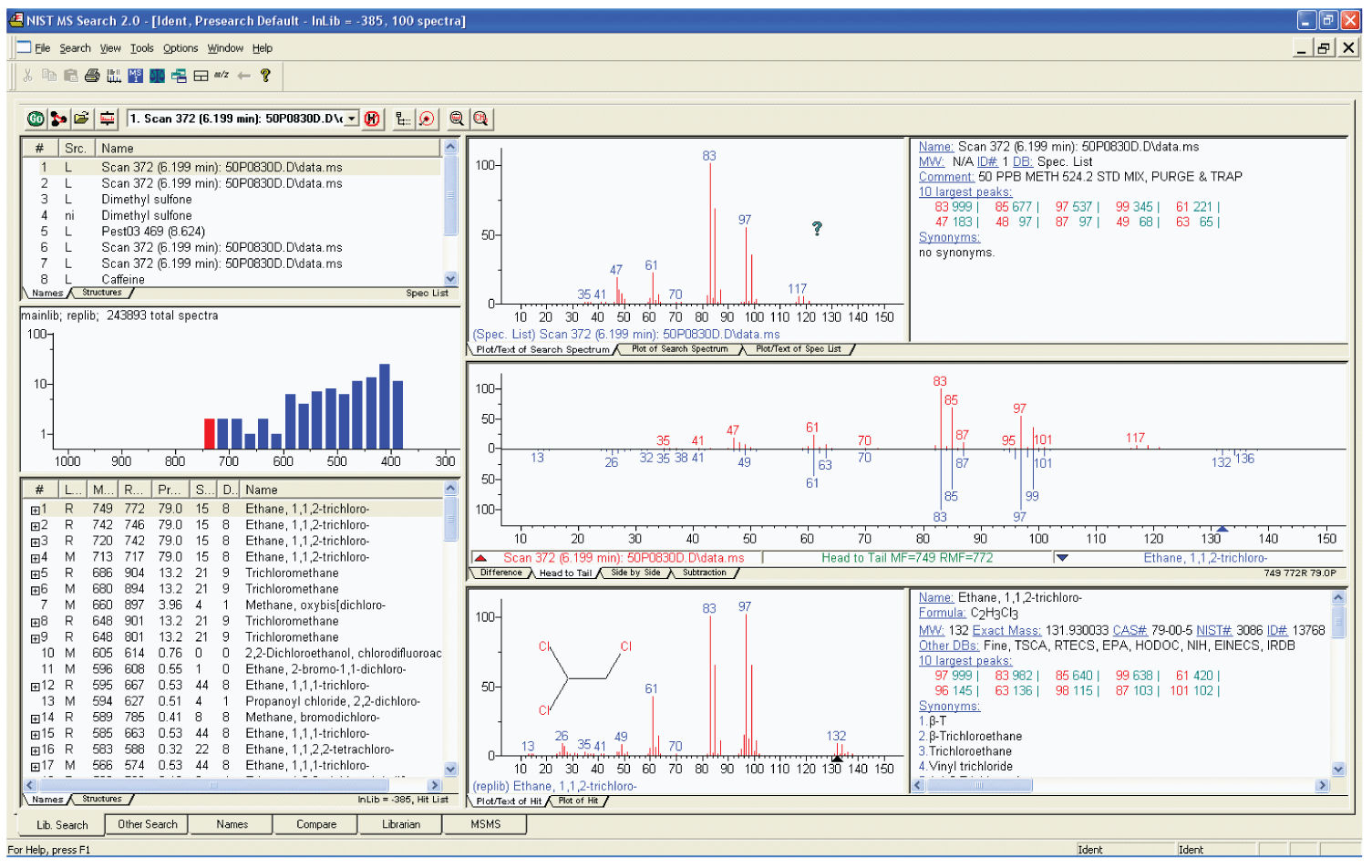 Figure 1: The Lib Search tab view of the NIST MS Search Program, called the Program’s Desktop. Other views are selected by clicking on the tabs on the lower left of the display. The Hit List can be displayed along with the hit number, database where the Hit is, Match Factor, Reverse Match factor, Probability, number of synonyms, and number of other databases containing the compound.
Figure 1: The Lib Search tab view of the NIST MS Search Program, called the Program’s Desktop. Other views are selected by clicking on the tabs on the lower left of the display. The Hit List can be displayed along with the hit number, database where the Hit is, Match Factor, Reverse Match factor, Probability, number of synonyms, and number of other databases containing the compound.
The Lib Search Tab Display
The Lib Search Tab is used to search imported spectra or structures against the NIST Mass Spectral Databases and other databases such the Wiley Registry of Mass Spectral Data and proprietary databases such as those that may be created by the user. Several different types of searches are available. Some are briefly described below. For a more detailed explanation, the NIST MS Search Program’s manual can be downloaded from http://chemdata.nist.gov/mass-spc/ms-search/docs/Ver20Man_11.pdf.
Spectra can be exported from Perkin Elmer’s TurboMass®, Waters’ MassLynx®, ThermoFisher’s Xcalibur®, Agilents’ GC GC/MS ChemStation®, LC LC/MS ChemStation®, and Mass Hunter®, JEOL/Sharder’s TSSPro®, and Bruker’s Workstation 6.9 to MS Search , brining it into focus, automatically searched, output the search results and return the focus to the calling program. For data systems that don’t have this connectivity, spectra in the mzML, netCDF, or JCAMP formats can be imported. NIST 11 is provided with AMDIS (Automated Mass spectral Deconvolution and Identification System) which reads the native form of many manufacturer’s GC/MS and LC/MS data file formats allowing for display of reconstructed total ion current (RTIC) chromatograms and selection of individual spectra to be sent to MS Search .
MS Search has several different search algorithms. The two primary categories are the Identity Search, which is used to determine if there is a mass spectrum of compound that generated the spectrum of the unknown in the Database, and the Similarity Search, which is used when it is believed that there is not a spectrum of the unknown in the Database and spectra of similar compounds to the unknown are retrieved as an aid to determining the identity of a unique substance. Both of these searches are variations of the NIST Mass Spectral Search algorithm which is describe in detail in the manual sited above. A spectrum can be search so that the best spectral matches are reported or the best spectral matches within a subset of conditions (constraints*) are reported. A spectrum search can be constrained as to what elements are presence and the number of atoms each element, the nominal mass range that the analyte may have, name fragments that may be present, name fragments that should not be present, specific elemental compositions, and the presences of peaks with specific m/z values and the relative intensity (ranges) of these peaks. These m/z -intensity pairs can be designated as normal peaks, peaks that represent losses from the molecular or precursor ion, or that represent a position in an intensity range; i.e., the most intense peak in the spectrum or the 2nd, 5th, etc. most intense. The most significant high m/z value peak can also be specified along with its intensity range.
New to v.2.0g is the ability to constrain a spectrum search by specifying an accurate mass of the analyte molecule along with a +/- accuracy range in parts-per-million (ppm), or millimass units (mmu). A numeric value for the mass or m/z value can be entered, or an elemental composition can be specified. This is epically useful when data are acquired with a time-of-flight (TOF) or some other type of mass spectrometer that provides accurate masses. All the spectra (both EI and MS/MS) in the NIST 11 Database are indexed according to their exact** mass. Anytime a spectrum record contains an elemental composition, not only will its nominal*** mass be reported, but its exact mass will also appear in the header of the Text Information Window for that spectrum. Spectra in User Databases can be indexed according to their exact masses so that these spectra can then have the exact mass as a constraint when these databases are searched. This means, that third party mass spectral databases such as the Wiley Registry and Wiley’s boutique databases and the Robert Adams Terpene database can be searched using the exact mass constraint.
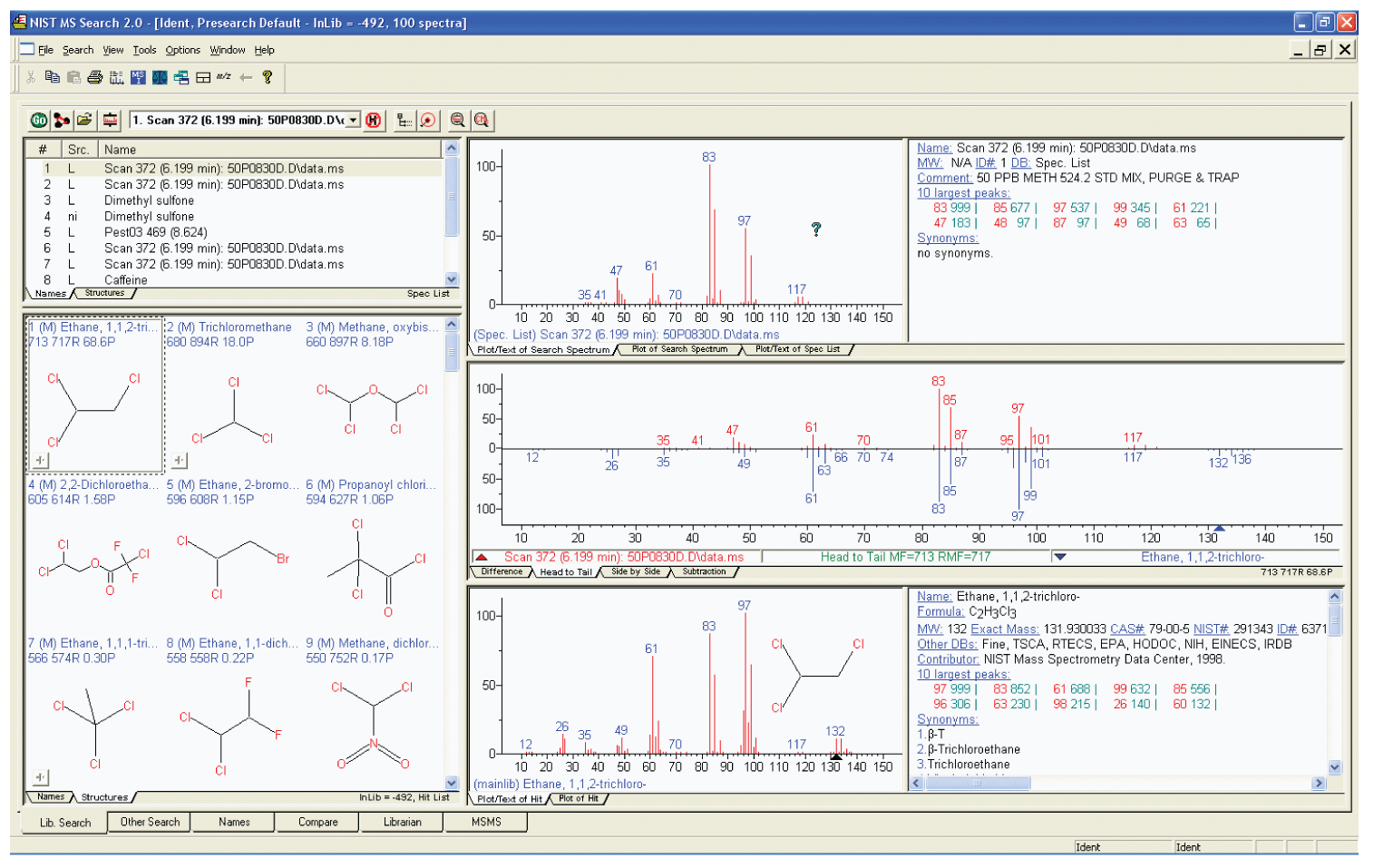
Figure 2: The Lib Search tab view with the Hit List displaying Hits as structures.
In addition to being able to search the Database of mass spectra using a submitted spectrum, it is also possible to perform a Structure Similarity Search. This allows for the submission of a structure that is imported from files using the molfile**** format or that exist on the Windows’ Clipboard in the molfile format and searching for similar structures in the Database resulting in a Hit List of compounds with similar structures. This is especially useful when dealing with an EI spectrum whose structure is suspected, but whose spectrum is believed not to be in the Database.
The features of the MS Search Program are designed to facilitate the identification of analytes. The first five and last column of the Hit List (lower left window in Figure 1) of Lib Search Tab display have to do with actual searches. Columns one and two are the Hit number and an Identification of which of the up to 127 simultaneously searched databases (New in v.2.0g; previous versions were limited to 16 databases) the Hit is in, respectively. The next three columns pertain to match of the searched spectrum to the library spectrum. The first of these columns is a Match Factor that uses a numerical scale of 0 to 999 based on both the m/z value and the intensities of all the peaks in both spectra. The second is a Reverse Match Factor which is calculated in the same way as the Match Factor; however, all peaks in the sample spectrum that are not in the library spectrum are disregarded when it is calculated. This is especially useful when the spectrum represent more than a single compound. The third value is a Probability, which is a more esoteric property of the Hit. It is not only dependent on the quality of the match, but also the uniqueness of the spectrum for that compound in the database compared to all the other spectra in the database. For example if spectra of the three regioisomers of xylene are in the Hit List, the probability numbers will not be high for these Hits due to the fact that their EI mass spectra are almost identical. The last column is the primary name of the compound that is assigned to spectra in the mainlib. Information from these six columns can be displayed with a display of the structure as seen in Figure 2 which is an alternative view of the Lib Search Tab.
Also New in v.2.0g of the MS Search Program is the optional display of the two columns between the Prob and Name columns. These columns show the number of synonyms associate with the Hit and the number of other database where the Hit’s compound can be found. These other databases are non-mass spectral databases. They include the Toxic Substance Control Act Inventory, EPA Environmental Monitoring Methods Index, European Index of Commercial Chemical Substances, and six others. When searching for known unknowns in various types of samples, two Hits which have similar numeric values can be differentiated from one another by the number of synonyms and the number of other databases containing the compound. The one with higher values for both of these is the more likely candidate. These two columns are also available in the Hit Lists produced in the Other Search tab view.
When a Lib Search tab Hit List is displayed it is usually sorted either by the Match Factor or the Reverse Match Factor, depending on the settings in the Library Search Options multiple tabbed dialog box.; however, by clicking on any of the column headers (except Lib.) the Hit List can be sorted by that column. Again, this is also true for the Hit List in Other Search tab view. The Spec List (upper left part of Figure 1) can be sorted alphabetically. This Spec List is the same as the Spec List in the Librarian tab view.
Some other import New features are the ability to have as many as 1,048,560 spectra in a single library instead of 786,420 spectra (especially important when trying to use the combined Wiley Registry/NIST Database ); import spectra from the Windows Clipboard that are in the NIST MSP text file format; ability to import spectra from mzXML and mzData MS and MS/MS files (in addition to mgf, msp, dta, pkl, JCAMP, etc.); optional display of isomers and derivatives as replicates (actually was introduced with v.20f); optionally turn off homologs in a Structure Similarity Search; and being able to do a string search in the Text Info Window. This last new feature is especially important when wanting to obtain information regarding a GC method associated with a Hit. If multiple methods are displayed, this feature can be used to find a specific column type or, especially because the journal article titles are included, the analysis of a substance in specific matrix.
It should not go unnoticed that v.2.0g is compatible with the latest version of the NIST Peptide MS/MS Databases . These are twelve free databases, (downloadable from http://peptide.nist.gov/) that contain >700K spectra (Human, Mouse, Drosophila, C. elegans, Yeats, E. coli, Rat, Chicken, Sigmaups1, and BSA). Just like the spectra of di- and tri-peptides shown the NIST 11 MS/MS Database, these spectra will optional be labeled with m/z values of peaks or ion types (b and y).
Covered in Parts 1 and 2 of this multiple part presentation on NIST 11, is the history of the NIST Mass Spectral Database(s), information about the search of spectra and structures and what is contained in the results of those searched, and the compatibility of with the NIST Peptides MS/MS Databases and where to find them. To be presented in subsequent parts are the uses of the searches in the Other Search tab view, how to use the NIST EI Database as an aid to determining the structure from a MS/MS spectrum without a match in the NIST MS/MS Database (includes Substructure Identification utility), the used the MS Interpreter (a tool that correlates structure to spectra and predicts spectra from structures), building user libraries with customized fields (Librarian tab view), and the Compare tab view.
* All constraints are not necessarily available at the same time. For example if an elemental composition is used as a constraints, the number of atoms of a specific element and/or the elements presence constraints are not available.
** An accurate mass is a value that is measured using a mass spectrometer. An exact mass is calculated form the published exact masses of the elements and their number in an elemental composition.
*** The nominal mass of an elemental is the integer mass of the most abundant naturally occurring isotope of that element. The nominal mass of a molecule, ion, or radical is calculated from the nominal masses of the elements in the elemental composition.
**** The Molfile is a file format that was created by Molecular Design Limited, founded in Hayward, CA in 1978 as a computer aided drug design firm. The company name was changed to Symyx until it merged with Accelrys. This ASCII file format contains information about the atoms, bonds, connectivity and coordinates of a molecule. The molfile consists of some header information, the Connection Table (CT) containing atom information, then bond connections and types, followed by sections for more complex information. It is considered an industry standard for chemical structures.




