Structural Identification of Unknowns from Spectra Obtained Using LC/MS and CAD Techniques and EI Mass Spectral Database.
This is the last in the five-part series about the features of NIST 11 (National Institute of Standards and Technology). This installment shows how spectra obtained by MS/MS methods from various soft ionization techniques, including those used with LC/MS can be searched against the NIST/EPA/NIH EI Mass Spectral Database and the results used to facilitate a structure determination.
When an organic ion fragments in the gas phase, the result is an ion and a neutral species. The fragment ion’s mass is recorded as part of the data record as a peak of a specific intensity, which is a function of the ion’s abundance, and at a specific m/z value, which is a function of its mass and the resolving power of the mass spectrometer. The mass of the neutral species (the dark matter of the mass spectrum) is inferred by the difference between the mass of the fragment ion and its precursor. The mass of both the ion and the dark matter are important in the determination of the structure of the compound that produced the mass spectrum. Oftentimes, the dark matter is more significant because it represents specific small groupings of atoms that are characteristic of a portion of the analyte. These neutral losses can often be attributed to specific substructuresa.
As the National Institute of Standards and Technology’s Mass Spectral Data Center began to develop ways by which its collection of electron ionization (EI) mass spectra could be searched, it became obvious that a search that allowed the dark matter (neutral losses) of an unknown spectrum to be searched against the dark matter of the spectra in the Database could be of great value in the identification of analytes whose mass spectra were not in the Database. Therefore, the spectra in the NIST/EPA/NIH Database were indexed as to their neutral losses as well as the m/z values of the peaks present. A mass spectrum (Demeton S-sulfone, an insecticide used to control aphids and other sucking insects, sawflies and spider mites on a range of crops) might exhibit a molecular ion peak at m/z 290. The peak at the next lowest m/z value that did not represent an ion containing a heavier mass isotope of one the elements or an ion that could be attributed to background was observed to have an m/z value of 197. Both of these values would be indexed for this compound in the Database. The difference in these two m/z values (93) which represented a neutral loss would also be indexed. This neutral loss represents the loss of a C2H5SO2 radical. The compound will also be indexed by this substructure and its substructures of an ethyl moiety, an SO2 functionality, and the presences of the heteroatoms O and S. The NIST/EPA/NIH Mass Spectral Database is currently indexed to a collection of 541 substructures [1]. Spectra with similar characterizing dark matter should have the same substructures. Identification of these substructures provides information about the pieces of the analyte that when fitted together can provide its structure because, “Mass spectrometry deals with the mass of the molecule and the mass of the pieces [substructures] of the molecule.” [2]
Dark-matter searching and identification of substructures is independent of fragmentation mechanisms and driving forces for the ion to break apart, and less dependent on relative peak intensity than of spectral-comparison searching. Therefore, the identification of substructures should be independent of the type of ion that fragments (molecular ion or protonated or diprotonated molecule). Searching of a mass spectrum of a protonated molecule fragmented through collisionally activated dissociation (CAD) against the dark matter of the NIST/EPA/NIH Mass Spectral Database can produce the same sub-structural information as would be produced if both the spectrum of the unknown and the spectra of the Database were produced by the same ionization techniqueb.
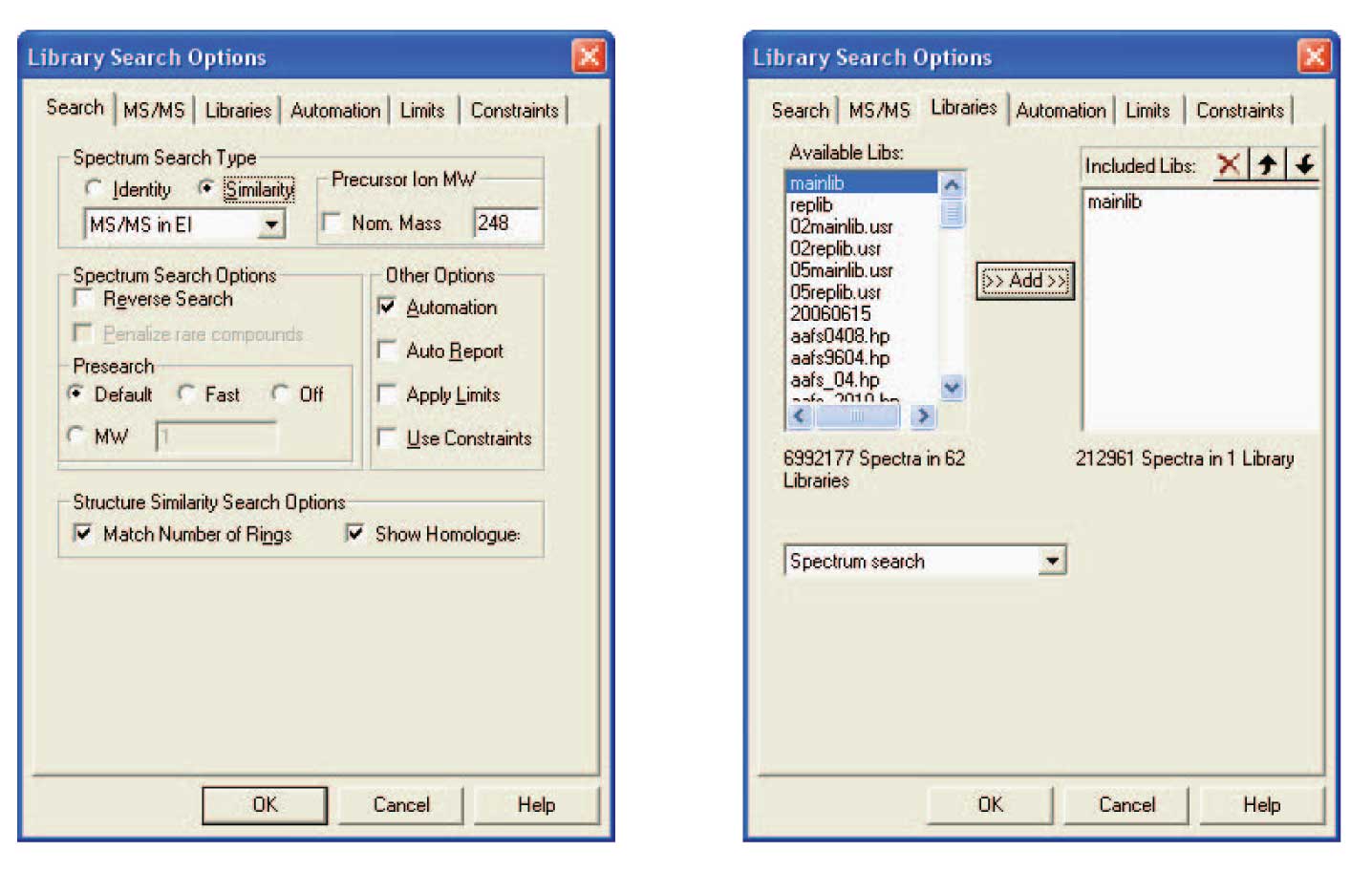
Figure 1: Left side is the Search Tab view of the Library Search Options Search showing the setup to perform a search of the EI Database to produce a Hit List which can be used by the Substructure Identification Tool. The right side of this figure is the Libraries Tab view showing that only the NIST mainlib Database of EI spectra has been selected.
The NIST Mass Spectral Search Program has a utility that examines the Hit List from a Spectrum Search against the NIST/EPA/NIH Database to determine the probability of the presences and absences of substructures. A special search algorithm was developed to produce a Hit List that is optimized for the Substructure Identification Tool . A spectrum obtained by MS/MS or in-source CAD from ions produced by APCI or electrospray is imported into the MS Search Program ’s Spec List. Using the Library Search Options Search dialog box’s Search tab view the Similarity search is selected, the MS/MS in EI option is chosen from the drop down list box, the Nom. Mass check box is deselected, and the m/z value of the appropriate precursor ions is entered (Figure 1). Then from the Libraries tab view make sure that the only selected database is the mainlib . Select the OK button to close the dialog box and store all the entered information. Highlight the imported spectrum in Spec List, and then click on the GO button; first button on the left of the button bar. The spectra in the Hit List may or may not resemble the spectrum that was searched; however, the spectra in the Hit List will have substructures that are in common or similar to those in the molecule that produced the CAD spectrum. After the search is complete, select Tools from the Main Menu bar at the top of the display. Select Substructure Identification from the Tools menu. This results in the display of the dialog box shown in Figure 2 which has a column of probable substructures that are present and a column of probable substructures that are absent.
In order to understand this concept, the term substructure needs to be defined as it pertains to a specific molecule. A substructure can be a heteroatom (N, O, S, Si, P, or a halogen), the number of rings and/or double bonds present, or a specific functionality (carbonyl, methyl ester, phenyl group, methyl group, etc.). The NIST substructure classification gets more specific than this with substructures such as ArO (an aromatic ring attached to an oxygen atom). Figure 3 illustrates some of the substructures associated with Meperidine (1-methyl-4-phenyl-4-piperidinecarboxylic acid, ethyl ester), the analyte used for illustration in this presentation.
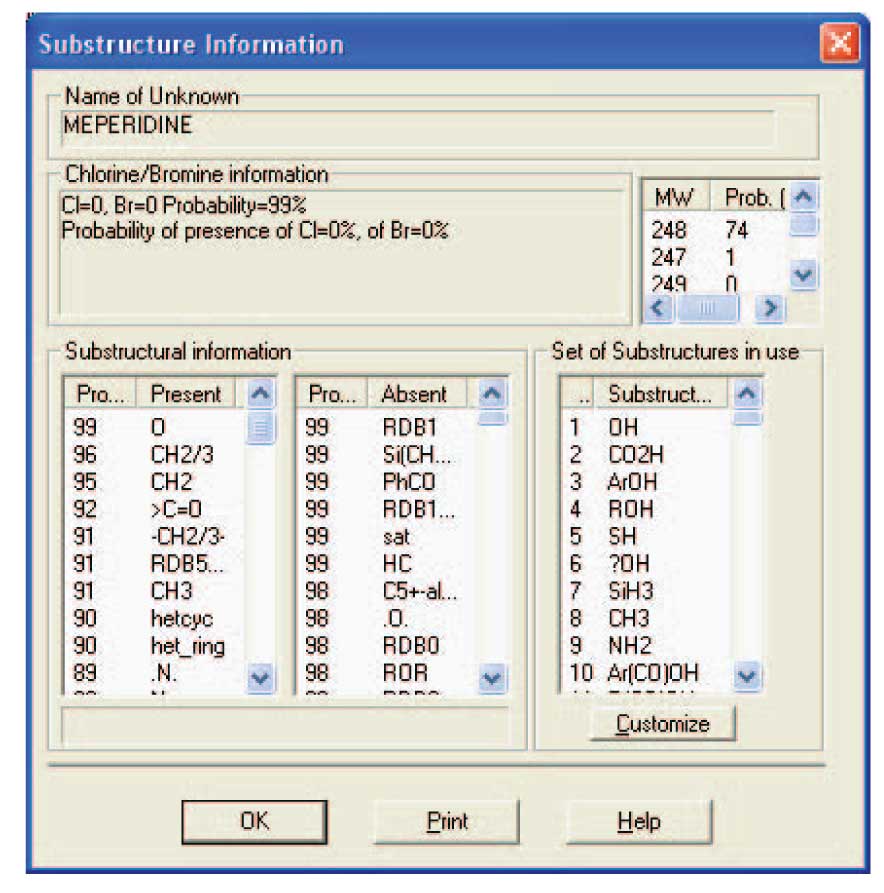
Figure 2: Substructure Information dialog box containing the results of the eveluation of a Hit List generated by an MS/MS in EI Similarity Search with m/z 248 specified as the precursor ion.
The mass spectrum of Meperidine (a controlled substance used as an analgesic in humans and also as a sedative anesthetic in animals) obtained by CAD (bottom of Figure 4) using the protonated molecule at m/z 248 as the precursor ion is used to illustrate the results that can be obtained from an MS/MS in EI option Similarity Search and substructure identification. After the search of the CAD spectrum against the NIST 11 Database of ~213K EI spectra is performed, a Hit List with 100 spectra is displayed. Whereas the spectrum of the Meperidine was obtained by CAD and the spectra in the Database were obtained by EI, the similarity in the compounds represented in the Hit List and the spectrum of the searched compound are of no relevance. What is relevant is the ability to predict the presence and the ability to predict the absence of substructures based on the substructures indexed to the spectra in the Hit List .
Examination of the Substructure Information dialog box from the NIST MS Search Program (top of Figure 4) shows that not only are the major substructures identified in the Probability Present column; but important information is also found in the Probability Absent column such as the fact that even though there is a high probability that an aromatic ring and an atom of oxygen are present, there is also a high probability that there is not an oxygen linked to an aromatic ring. These types of data can lead to elucidation of the analyte’s structure.
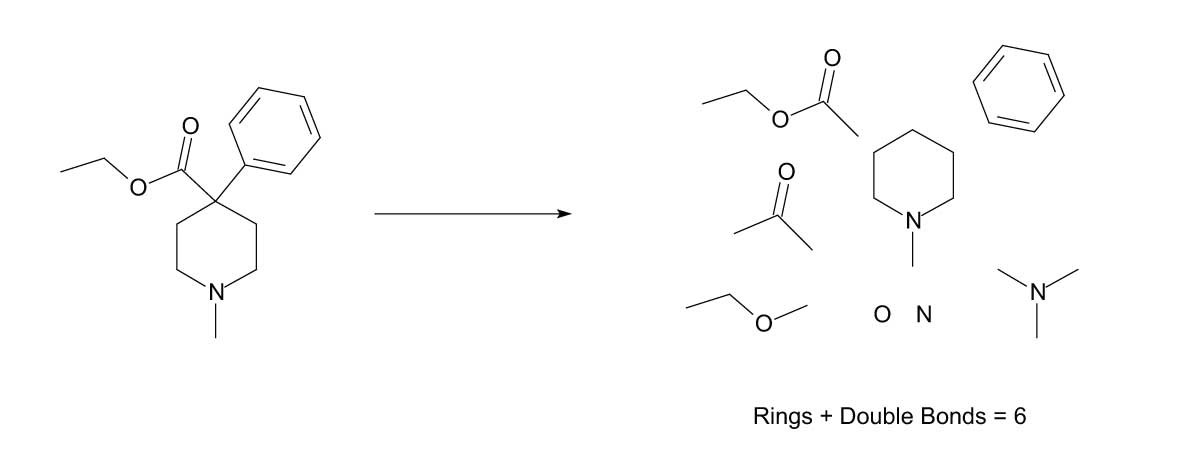 Figure 3: Examples of some of the substructures that may be considered for Meperidine.
Figure 3: Examples of some of the substructures that may be considered for Meperidine.
The Substructure Identification tool within the NIST MS Search Program has been enhanced and modified in the NIST 11 release. As is shown in this presentation, the substructure information that was extractable from the MS/MS in EI Similarity Search Hit List produces higher probabilities of substructures present and more substructures than those shown in Figure 3. It is important to emphasize the value of the Probability Absent values. These values are calculated and not just a difference from the Probability Present list.
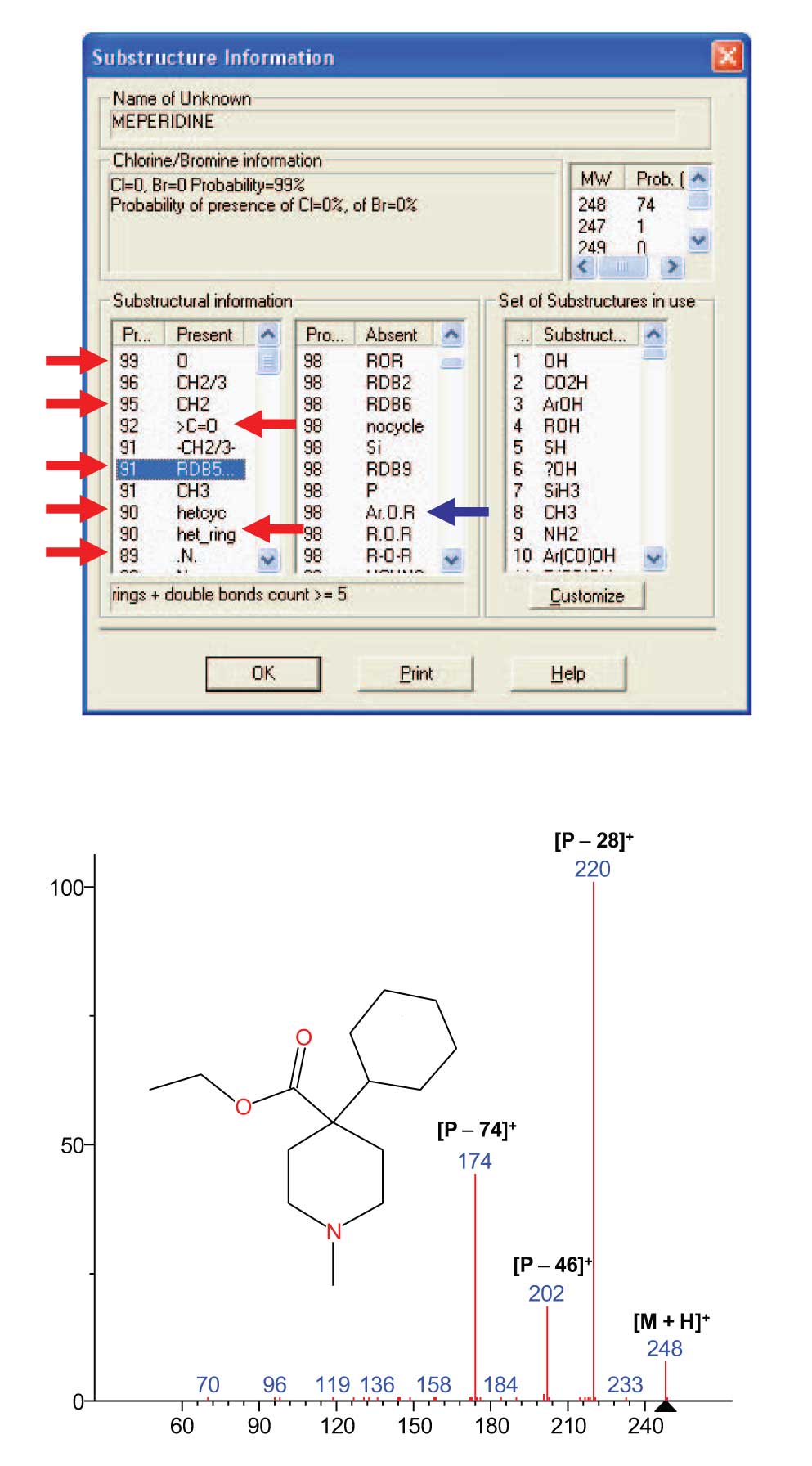 Figure 4: (Top) Results of the Substructure Identification Tool applied to the Hit List obtained from a an MS/MS in EI Similarity Search of the NIST 11 EI Database of the mass spectrum acquired using CAD of the protonated molecule of Meperidine resulting from electrospray ionization (Bottom).
Figure 4: (Top) Results of the Substructure Identification Tool applied to the Hit List obtained from a an MS/MS in EI Similarity Search of the NIST 11 EI Database of the mass spectrum acquired using CAD of the protonated molecule of Meperidine resulting from electrospray ionization (Bottom).
Currently, the spectra in the NIST MS/MS databases are being evaluated with respect to information that can be provided using the MS/MS in EI Similarity Search and the extraction of substructure information from the resulting Hit List.
It can no longer be said that NIST/EPA/NIH Mass Spectral Database is an EI Database only and that the NIST Mass Spectral Search Program is only for use with GC/MS EI data. As seen in this presentation, the EI Database has a great deal of utility in the determination of structures from CAD data obtained on ions produced by LC/MS. As seen in earlier presentations in this series, the NIST MS/MS database can be used to identify spectra produced by CAD process of ions that originate with LC/MS. There are other ways to use the NIST Database of mass spectra (both the EI and MS/MS databases) with LC/MS data. For example, if an elemental composition of an ion can be determined from an accurate mass measured on an LC/MS instrument, that elemental composition can be searched against the NIST EI database to see what compounds with that elemental composition are present. Using the number of synonyms and/or presence in other databases (non-mass spectral database) can aid in choosing which of the many compounds is a good candidate. Accurate masses obtained by various LC/MS instruments can also be used in a similar way. The number of ways that the NIST Databases and MS Search Program is only limited by imagination.
This is the final installment in this series. Look for future articles on AMDIS and other aspects of mass spectrometry.
a. A substructure is a characterizing part or attribute of an ion. It can be a functionality; i.e., an ethyl group, a carbonyl, or double or triple bond, the presence and/or position a heteroatom, the number of rings and/or double bonds, or combinations of such attributes.
b. It should be noted that the NIST/EPA/NIH EI Mass Spectral Database is the only commercially available database that is indexed according to substructures. If the search is done against another database such as the Wiley Registry substructure information cannot be extracted.
References:
[1] Stein, SE “Chemical Substructure Identification by Mass Spectral Searching” J. Am. Soc. Mass Spectrom. 1995, 6, 644–655.
[2] McLafferty, FW :Computer Mass Spectrometry Training” Agilent Technologies, 1989.
[3] Baumann, C; Cintora, MA; Eichler, M; Lifante, E; Cooke, M; Przyborowska, A; Halket, JM “A Library of Atmospheric Pressure Ionization Daughter Ion Mass Spectra Based on Wideband Excitation in an Ion Trap Mass Spectrometer” Rapid Comm. Mass Spectrom. 2000,14(5), 349–356.
O. David Sparkman is currently an Adjunct Professor of Chemistry at the University of the Pacific in Stockton, California; Contractor to the National Institute of Standards and Technology Mass Spectrometry Data Center; President of ChemUserWorld.com; and a former American Chemical Society Instructor (1978–2006) and American Society for Mass Spectrometry Member-at-large for Education (2004–2006).
At the University of the Pacific, Prof. Sparkman teaches courses in mass spectrometry and analytical chemistry and manages the mass spectrometry facility. Over the past 28 years, he has developed and taught five different ACS courses in mass spectrometry; he holds positions on the Editorial Advisory Boards of the European Journal of Mass Spectrometry and the HD Science GC/MS Update – Part B; and is the Book Review Editor for the European Journal of Mass Spectrometry. He is the author of Mass Spectrometry Desk Reference (Global View Publishing: Pittsburgh, PA, 1st ed. 2000; 2nd ed. 2006). Prof. Sparkman is a member of the Editorial Boards of the John Wiley Encyclopedia of Environmental Analysis and Remediation and Encyclopedia of Analytical Chemistry, Editor of and a contributor to the Mass Spectrometry Section of the Encyclopedia of Analytical Chemistry, and a contributor to the Encyclopedia of Environmental Analysis and Remediation. Along with J. Throck Watson, he developed the Mass Spectral Interpretation Quick Reference Guide.
He also provides general consulting service in mass spectrometry for a number of instrument manufacturers, manufacturing companies, and government agencies.




